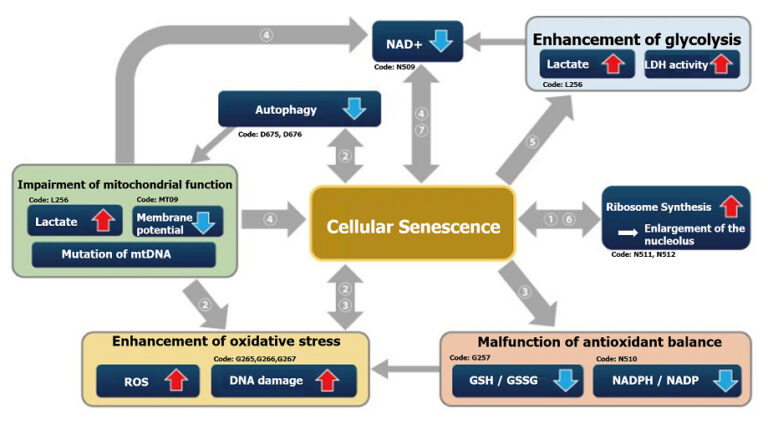
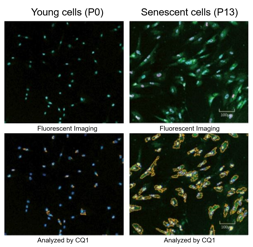
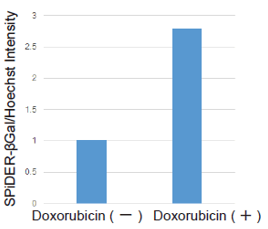
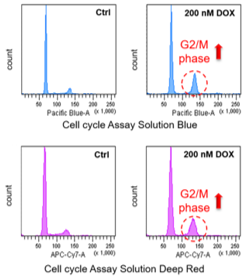
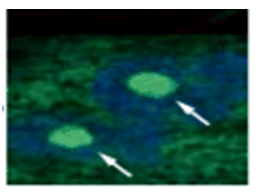
| |
Cell |
Senescence induction |
Senescence marker (s) |
Responding variable (s) |
Reference |
| ① |
IMR90
(Human pulmonary fibroblasts) |
Several passages in culture |
SA-ß-Gal, p16, p21, Nucleosome hypertrophy |
Expression of SETD8↓, H4K20me1↓, oxidative phosphorylation↑, ribosome synthesis↑ |
H. Tanaka, S. Takebayashi, A. Sakamoto, N. Saitoh, S. Hino and M. Nakao, “The SETD8/PR-Set7 Methyltransferase Functions as a Barrier to Prevent Senescence-Associated Metabolic Remodeling.”, Cell Reports, 2017, 18(9), 2148. |
Inhibition of SETD8
(Methyltransferase) |
Oxidative phosphorylation↑, ribosome synthesis↑ |
| ② |
Senescent mouse satellite cell
eletal muscle progenitor cells) |
– |
SA-ß-Gal, p16 |
Autophagy activity↓, ROS↑, mitochondrial membrane potential↓ |
L. Garcia-Prat, M. Martinez-Vicente and P. Munoz-Canoves, “Autophagy: a decisive process for stemness”, Oncotarget, 2016, 7(11), 12286. |
Atg7 knockout mouse
(Satellite cells) |
Autophagy inhibition |
SA-ß-Gal, P15, p16, p21, γ-H2AX |
ROS↑, mitochondrial membrane potential↓ |
| ③ |
Rat fibroblast model of type 2 diabetes |
– |
SA-ß-Gal, p21, p53, γ-H2AX |
NADP+/ NADPH↓(resistance to oxidative stress↓), NADPH oxidase↑(ROS↑) |
M. Bitar, S. Abdel-Halim and F. Al-Mulla, “Caveolin-1/PTRF upregulation constitutes a mechanism for mediating p53-induced cellular senescence: implications for evidence-based therapy of delayed wound healing in diabetes”, Am J Physiol Endocrinol Metab., 2013, 305(8), E951. |
| ④ |
IMR90
(Human pulmonary fibroblasts) |
Ethidium bromide (inhibition of mtDNA) + pyruvate deficiency |
SA-ß-Gal |
NAD+/NADH |
C. Wiley, M. Velarde, P. Lecot, A. Gerencser, E. Verdin, J. Campisi, et. al., “Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype”, Cell Metab., 2016, 23(2), 303. |
| ⑤ |
MDA-MB-231
(Human breast cancer cells) |
X-ray irradiation + inhibition of cell cycle-related factor (securin) expression |
SA-ß-Gal |
Lactate↑, LDH activity↑, (glycolysis↑) |
E. Liao, Y. Hsu, Q. Chuah, Y. Lee, J. Hu, T. Huang, P-M Yang & S-J Chiu, “Radiation induces senescence and a bystander effect through metabolic alterations.”, Cell Death Dis., 2014, 5, e1255. |
| ⑥ |
MEF
(Mouse Embryonic Fibroblast) |
Overexpression of oncogenes,several passages in culture, transcription factor overexpression(E2F1) |
SA-ß-Gal, p16, p21, Nucleosome hypertrophy |
Ribosome RNA↑, p53↑ |
K. Nishimura, T. Kumazawa, T. Kuroda, A. Murayama, J. Yanagisawa and K. Kimura, “Perturbation of Ribosome Biogenesis Drives Cells into Senescence through 5S RNP-Mediated p53 Activation”, Cell Rep. 2015, 10(8), 1310. |
| ⑦ |
Mouse tail fibroblast |
2 months old, 22 months old, p16 knockout (22 months old) |
SA-ß-Gal, p14, p16 |
NAD+↓, SIRT3↓ |
M. J. Son, Y. Kwon, T. Son and Y. S. Cho, “Restoration of Mitochondrial NAD+ Levels Delays Stem Cell Senescence and Facilitates Reprogramming of Aged Somatic Cells”, Stem Cells. 2016, 34(12), 2840. |